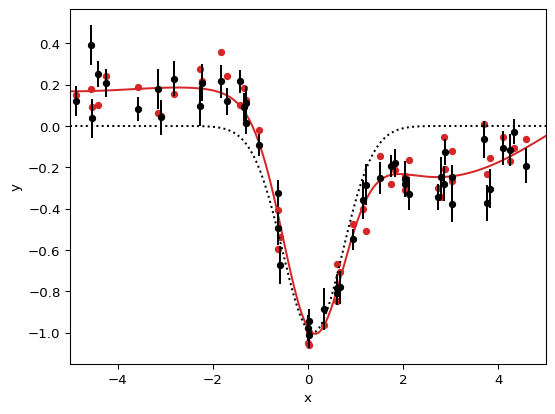
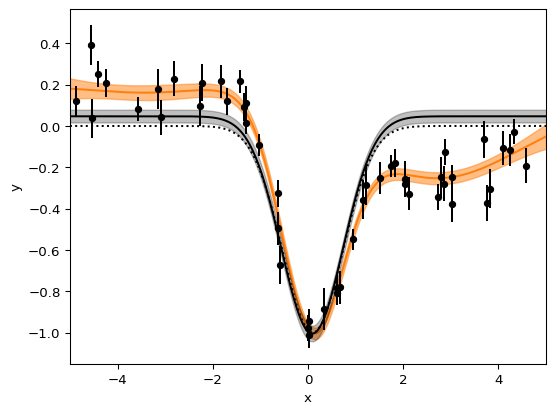
csv = """
x,y,y_err
-4.862316,0.119011,0.072891
-4.566759,0.39243,0.09602
-4.526447,0.03616,0.093953
-4.401908,0.252323,0.062631
-4.246188,0.205874,0.0674
-3.562332,0.080907,0.059129
-3.157129,0.178284,0.09509
-3.084805,0.041965,0.085326
-2.812079,0.229085,0.086333
-2.274074,0.096489,0.095004
-2.235357,0.209088,0.088958
-1.831639,0.216567,0.079958
-1.703316,0.119445,0.064556
-1.421827,0.215532,0.05757
-1.35114,0.092736,0.066759
-1.31176,0.112926,0.082878
-1.297492,0.016536,0.053667
-1.027974,-0.09196,0.05275
-0.638266,-0.325933,0.06616
-0.622723,-0.495434,0.079524
-0.578592,-0.673282,0.092695
0.009951,-0.977624,0.064353
0.029668,-1.014229,0.058653
0.030832,-0.9436,0.056701
0.333102,-0.885644,0.099733
0.611962,-0.80845,0.058975
0.614331,-0.783845,0.065877
0.680987,-0.781937,0.078415
0.946248,-0.54763,0.050467
1.153962,-0.356636,0.095032
1.221088,-0.284847,0.098862
1.513781,-0.250288,0.077845
1.748809,-0.194554,0.054239
1.834629,-0.179785,0.06665
2.04261,-0.25717,0.086421
2.045813,-0.282465,0.057122
2.12702,-0.326968,0.077623
2.728266,-0.342656,0.063652
2.799758,-0.24829,0.098725
2.853586,-0.278951,0.083389
2.887301,-0.125747,0.062783
3.018722,-0.245938,0.055416
3.021476,-0.377726,0.088809
3.691274,-0.063975,0.089124
3.759326,-0.37162,0.08808
3.826412,-0.302928,0.09572
4.09316,-0.108121,0.082931
4.248676,-0.117814,0.078418
4.331401,-0.028635,0.060088
4.581394,-0.193597,0.084915
"""
data = pd.read_csv(io.StringIO(csv))