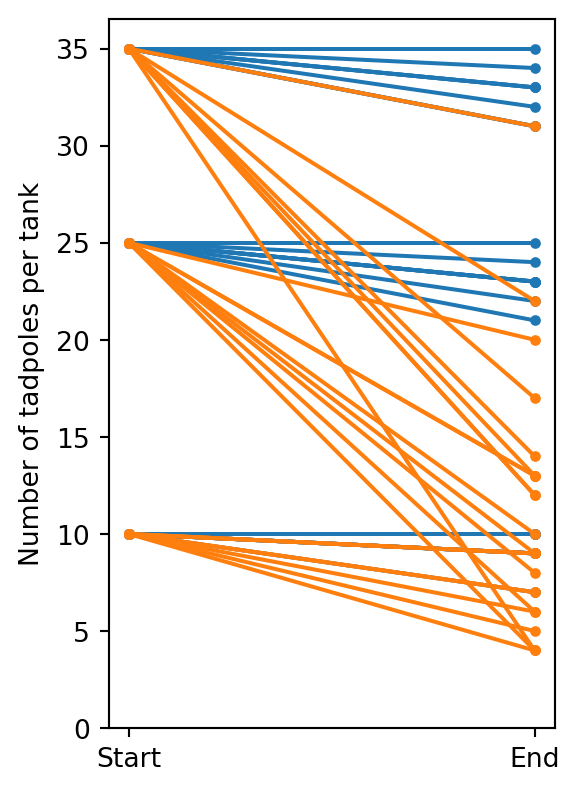
csv = """
num_start,has_predators,is_large,num_end
10,0,1,9
10,0,1,10
10,0,1,7
10,0,1,10
10,0,0,9
10,0,0,9
10,0,0,10
10,0,0,9
10,1,1,4
10,1,1,9
10,1,1,7
10,1,1,6
10,1,0,7
10,1,0,5
10,1,0,9
10,1,0,9
25,0,1,24
25,0,1,23
25,0,1,22
25,0,1,25
25,0,0,23
25,0,0,23
25,0,0,23
25,0,0,21
25,1,1,6
25,1,1,13
25,1,1,4
25,1,1,9
25,1,0,13
25,1,0,20
25,1,0,8
25,1,0,10
35,0,1,34
35,0,1,33
35,0,1,33
35,0,1,31
35,0,0,31
35,0,0,35
35,0,0,33
35,0,0,32
35,1,1,4
35,1,1,12
35,1,1,13
35,1,1,14
35,1,0,22
35,1,0,12
35,1,0,31
35,1,0,17
"""
data = pd.read_csv(io.StringIO(csv))
data = data.drop(columns=["is_large"])